Area-wide testing by high-throughput diagnostics
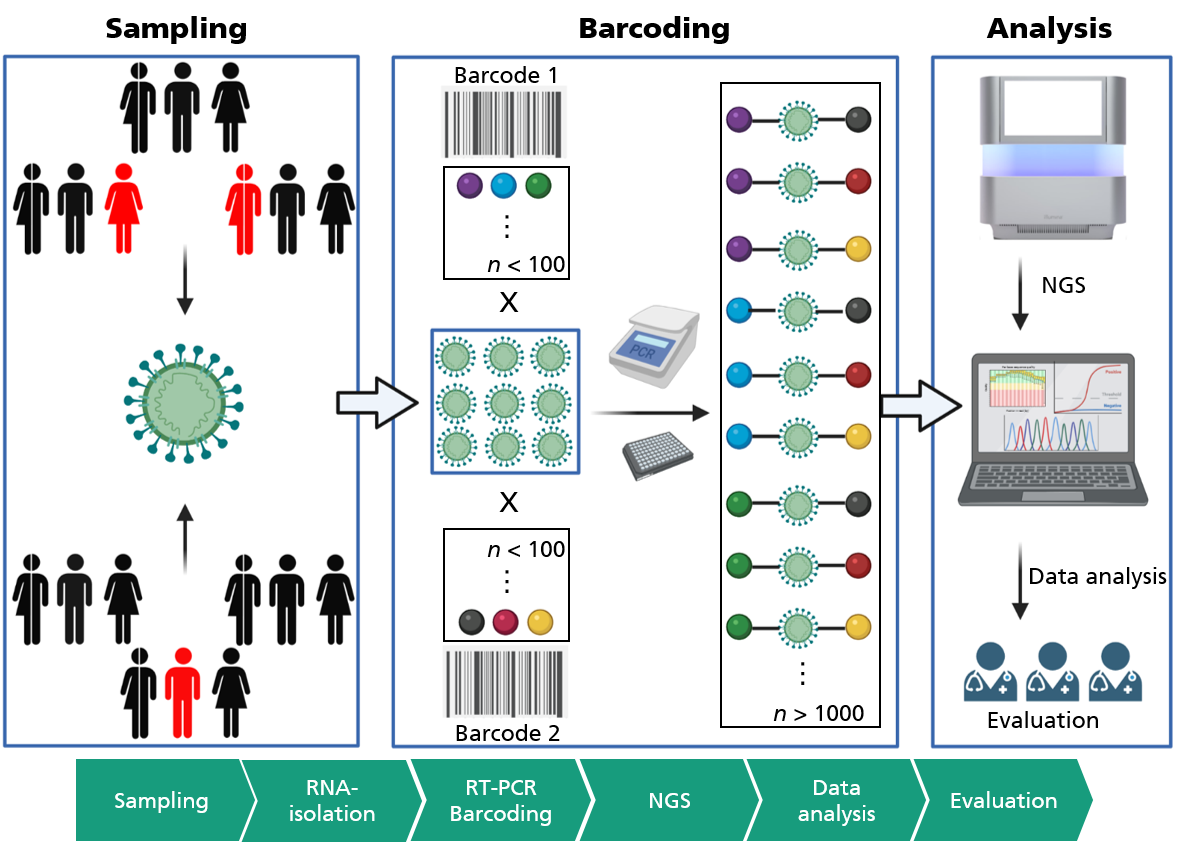
Therefore, in order to be able to offer area-wide PCR testing with regard to the current Omikron variant, sample throughput must be significantly increased. In collaboration with the Fraunhofer Institutes for Cell Therapy and Immunology IZI, and Production Technology and Automation IPA, we at Fraunhofer IGB have developed a new method for high-throughput diagnostics of SARS-CoV-2 and validated it on clinical samples as part of the Fraunhofer CoV-2-KomET project. This combines reverse transcription and amplification of specific viral sequences (RT-PCR) with the high-throughput capability of modern sequencing technologies. In order to analyze thousands of patients simultaneously, samples are labeled with specific oligonucleotides – also known as “molecular barcodes” – in the RT-PCR step, which allows a unique assignment to the patient (see Fig.).
First validation of the approach
CoV-2-KomET was successfully tested on a total of 672 clinical samples and achieved a specificity of over 89 percent and a sensitivity of over 87 percent in comparison to standard PCR tests. The diagnostic value of CoV-2-KomET, determined by the “area under the curve (AUC)”, of 94 percent also demonstrates the potential of this approach, which will be subject to further evaluation and improvement in the future. In addition to the pure SARS-CoV-2 high-throughput detection, a “respiratory panel” was successfully established on 41 synthetic samples, allowing the detection of SARS-CoV-2 as well as Influenza A/B down to a detection limit of 10 copies/µL.
Outlook
CoV-2-KoMET, as a diagnostic approach based on high-throughput sequencing of viral nucleic acids, represents a promising and alternative method for pandemics and infection outbreaks. Not only could sample throughput be increased with this approach, but scalability could also reduce the potential costs per test. Furthermore, by changing the design of the specific viral sequences, it is possible to respond to new variants within a few weeks. The project will be continued with the industrial partner LMV Laboratories, with a special focus on improving and expanding the respiratory panel to simultaneously test patients for SARS-CoV-2, Influenza and RSV within one sample.

 Fraunhofer Institute for Interfacial Engineering and Biotechnology IGB
Fraunhofer Institute for Interfacial Engineering and Biotechnology IGB