Sepsis is caused by infections of the bloodstream and is responsible for approximately 60,000 deaths every year in Germany alone. In order to quickly and reliably diagnose the pathogens that cause the disease, the innovation field is working with leading hospitals in Germany to develop and evaluate a diagnostic method based on high-throughput sequencing (NGS) of circulating nucleic acids in plasma.
NGS-based diagnostics of sepsis pathogens
Sepsis remains one of the main causes of death in intensive care units. It is caused by infections, whereby pathogens – bacteria, viruses, fungi or parasites – spread throughout the body from a local center of infection via the lymph and the bloodstream, triggering an uncontrolled and overreactive immune response. Rapid treatment with the appropriate antibiotic is critical for the patient’s survival. However, it often takes too long to reliably identify the corresponding sepsis pathogen. Furthermore, standard blood culture analysis – which uses microbiological methods to detect microbes – only manages to diagnose the original pathogen in less than 30 percent of cases.
Disadvantages of microbial methods of pathogen identification
Classical methods for pathogen identification usually are based on microbiological detection of the pathogen using so-called blood cultures with subsequent identification of the corresponding species using microscopic, biochemical or mass spectroscopic methods. Disadvantages of the microbiological approach are low detection rates and longer times required for an unambiguous diagnosis. In addition, the result is often negative even though the disease is actually caused by an infection, because some pathogens cannot be cultivated at all or only under laboratory conditions.
Our approach: NGS diagnosis of pathogens
For this reason, we develop innovative molecular methods for pathogen diagnostics on behalf of and in cooperation with customers and partners from industry and hospitals. These new methods are based on molecular analyses of the genetic information of pathogens. The three-stage process comprises optimal sample preparation, high-throughput sequencing (Next-Generation Sequencing, NGS) and bioinformatics analysis using proprietary diagnostic algorithms.
Advantage: Fast and reliable diagnosis
The new technology avoids time-consuming cultivation procedures and makes possible the detection of all pathogens: viruses, parasites and bacteria that do not grow on the culture media used. This not only makes the diagnosis faster, but also significantly more reliable. The NGS data can also be used for pathogen diagnostics, or for the identification of new biomarkers.
Application areas of NGS diagnostics
We have extensive experience in the indications of sepsis, endocarditis, amniotic fluid infections, as well as in biomarker screening, the genome characterization of pathogens and microbial studies (e.g. on the skin).
Thanks to its comprehensive, open platform character, this type of infection diagnostics already holds the potential for universal use in hospitals. With third-generation sequencers, immediate point-of-care diagnosis is also within reach, as we have been able to show in successful pilot studies.
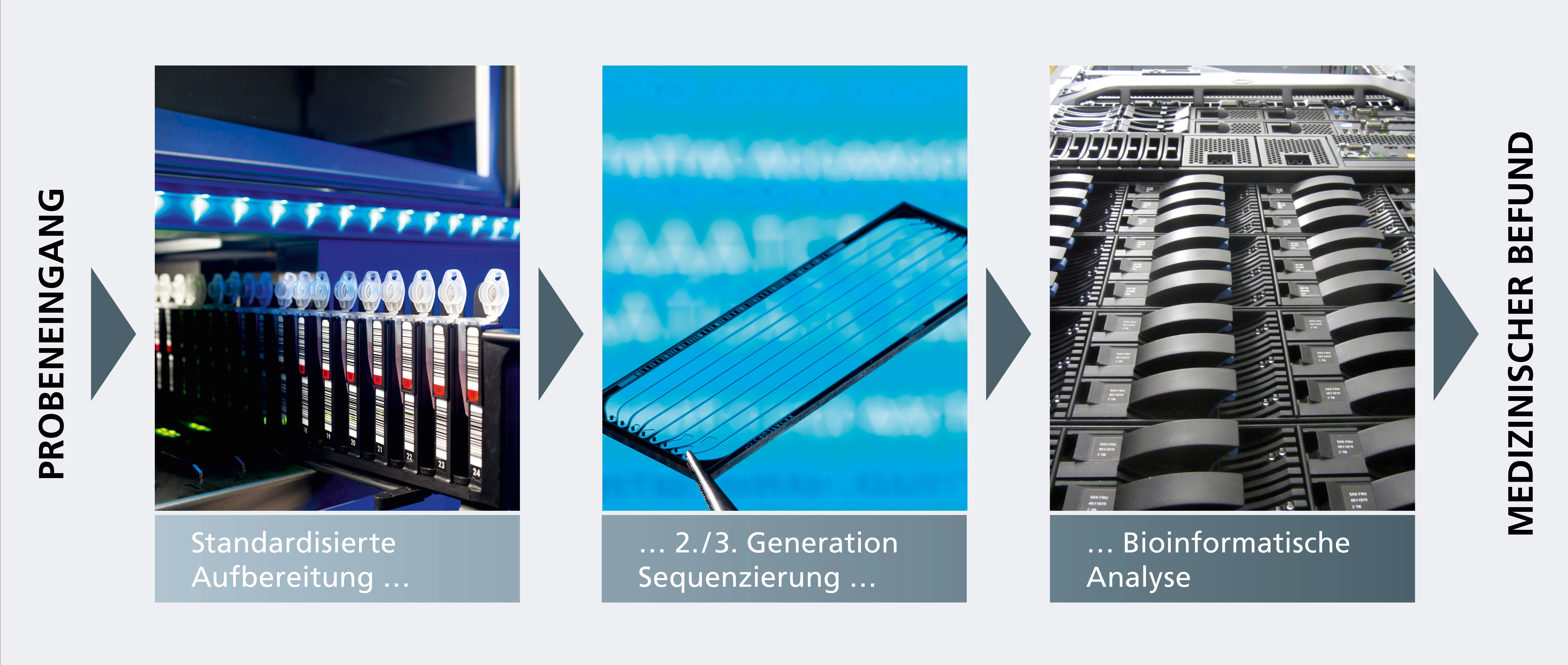
Next-generation diagnostics for sepsis pathogens
Here at Fraunhofer IGB, we have developed an innovative process that relies on molecular diagnosis and bioinformatics. Using next-generation sequencing (NGS) in combination with bioinformatic algorithms, this process is able to detect DNA fragments of pathogens in the patient’s blood to identify the cause of the sepsis with highest specificity and sensitivity [1–3].
Identification of pathogens using microbial, cell-free DNA
Our approach exploits the fact that the DNA sequences of every pathogen are unique and that cells release DNA in the form of circulating, cell-free fragments (cfDNA) – healthy cells, cancer cells and microorganisms that can be analyzed to identify pathogens. Comparable to liquid biopsy, cfDNA is isolated from blood using a fully automated method which is subsequently sequenced using NGS.
To identify the pathogens, the sequences of all the DNA fragments present within a blood sample – up to 30 million of them – are bioinformatically aligned with known human sequences stored in databases. This comparison reveals that less than one percent of the cfDNA originates from microbes rather than from the patient themselves. In the next step, we compare these “reads” precisely with the sequences that are stored in a specially developed database containing the genomes of bacteria, viruses, fungi and parasites.
Via a bioinformatic analysis, an SIQ (sepsis indicating quantifier) score is determined for each identified pathogen and compared to the score from non-infected controls to detect lowest levels of contamination. If the SIQ score exceeds a threshold value, the relevant patients are diagnosed as SIQ positive for the pathogen concerned [1].
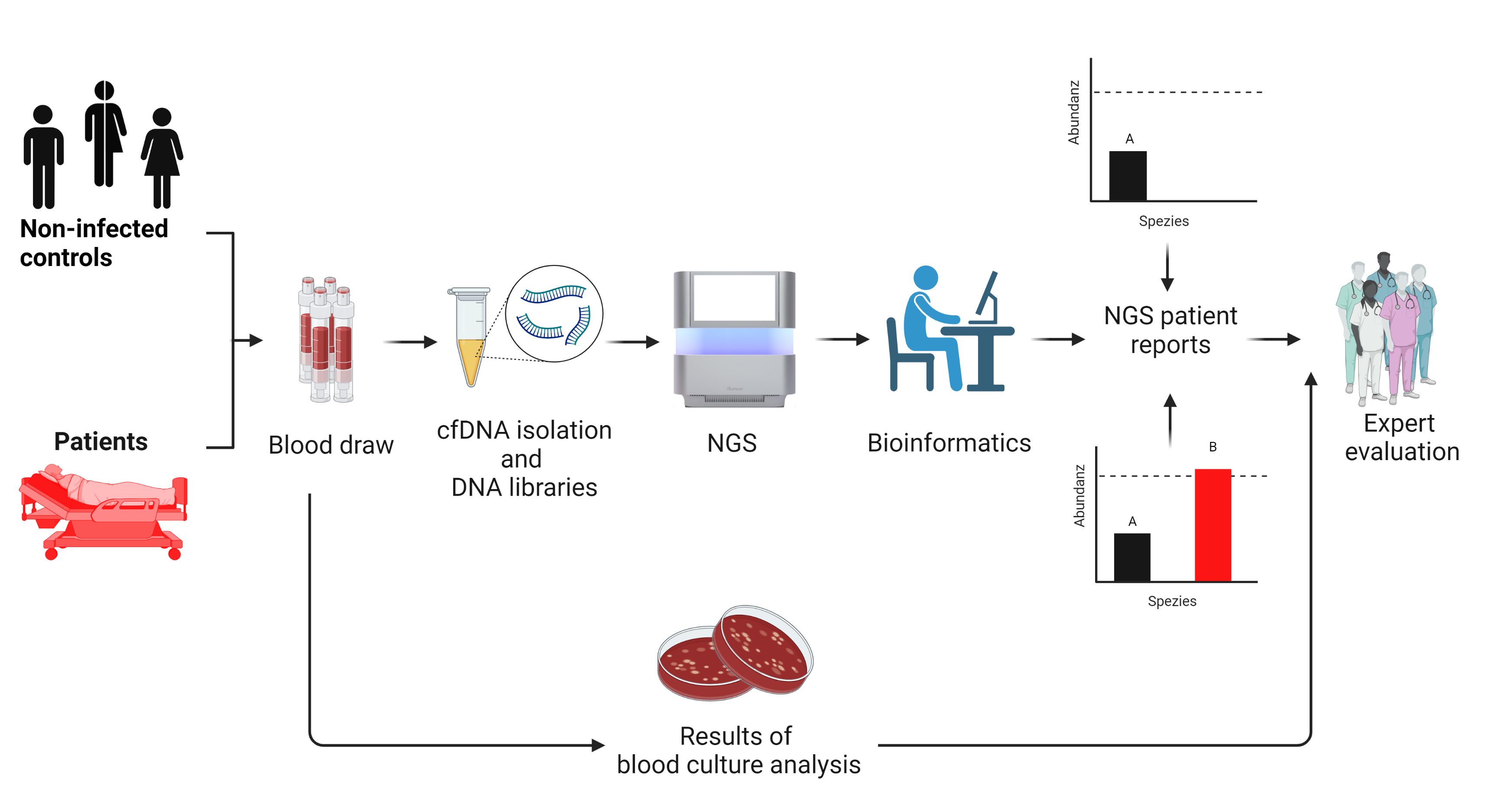
Clinical evaluation of NGS-based diagnostics of sepsis pathogens
Monocentric clinical study in collaboration with the University Hospital Heidelberg
As part of a recent clinical study in collaboration with the University Hospital Heidelberg, we were successful in achieving significantly more positive results on pathogen identification with this technology compared to using blood culture (71 percent compared to 11 percent) in 50 patients with septic shock [3]. A jury of independent intensive care medics considered 96 percent of the positive NGS results to be plausible.
According to the jury, treatment of 53 percent of the patients would have been adapted subsequently on the basis of these results, since they were often over- or undertreated due to the empirical antibiosis. In this group of patients not receiving adequate treatment, the mortality rate was increased by 13 percent [3].
These concrete effects on patient treatment success convincingly demonstrate the enormous potential of more reliable and sensitive pathogen diagnostics. The retrospective observations will now be validated in a multicenter study with approximately 15 hospitals in Germany.
Multicentric clinical study
To further validate these promising results, “Next GeneSiS” was launched in 2019 – a multicentric clinical study encompassing 500 sepsis patients and 50 healthy subjects at 17 clinics [5]. The clinical value of the NGS-based approach is being evaluated by a committee of independent clinical experts judging on plausibility and possible changes to the treatment of the patients based on the NGS results.
The study data has not yet been published but interim evaluations indicate that the previous results obtained for the NGS approach with regard to sensitivity, specificity and plausibility are now being corroborated in a large, independent patient population.
The Dietmar Hopp Stiftung (Dietmar Hopp Foundation) is sponsoring the multicentric study along with a research project at Essen and Heidelberg University Hospitals, where we are investigating NGS-based diagnosis on seriously ill newborn babies, premature infants and small children [6].
Infection diagnostics 3.0

Reliable and timely diagnosis of pathogens in critically ill patients still poses major problems for intensive care medicine. By establishing a diagnostic procedure based on high-throughput sequencing (NGS) of microbial DNA from circulation of patient blood, Fraunhofer IGB has already been able to establish a procedure for the reliable diagnosis of pathogens in the past, which has a detection rate five to six times better than that of culture-based procedures [1, 3].
Third generation single molecule DNA sequencing technologies for real-time point-of-care diagnostics
With the help of latest single molecule DNA sequencing technologies of the third generation, this method has now been further developed so that real-time analysis of microbial DNA is already possible during sequencing. Consequently, the identification of pathogens can be reduced to six to eight hours [4]. Thus, the reliability of sequencing-based diagnostics can be combined with the speed advantage of real-time analysis in order to provide patients with optimal antibiotic therapy as quickly as possible in the future.
References
[1] Grumaz, S.; Stevens, P.; Grumaz, C.; Decker, S.O.; Weigand, M.A.; Hofer, S.; Brenner, T.; von Haeseler, A.; Sohn, K. (2016) Next-generation sequencing diagnostics of bacteremia in septic patients, Genome Medicine 8:73, doi: 10.1186/s13073-016-0326-8
[2] Decker, S.O.; Sigl, A.; Grumaz, C.; Stevens, P.; Vainshtein, Y.; Zimmermann, S.; Weigand, M.A.; Hofer, S.; Sohn, K; Brenner, T. (2017) Immune-response patterns and next generation sequencing diagnostics for the detection of mycoses in patients with septic shock – results of a combined clinical and experimental investigation, International Journal of Molecular Sciences 18, 1796, doi: 10.3390/ijms18081796
[3] Grumaz, S.; Grumaz, C.; Vainshtein, Y.; Stevens, P.; Glanz, K.; Decker, S.O.; Hofer, S.; Weigand, M.A.; Brenner, T.; Sohn, K. (2019) Enhanced performance of next-generation sequencing diagnostics compared to standard of care microbiological diagnostics in patients suffering from septic shock, Critical Care Medicine, doi: 10.1097/CCM.0000000000003658
[4] Grumaz, C; Hoffmann, A.; Vainshtein, Y.; Kopp, M.; Grumaz, S.; Stevens, P.; Decker, S. O.; Weigand, M. A.; Hofer, S.; Brenner, T.; Sohn, K. (2020) Rapid next generation sequencing-based diagnostics of bacteremia in septic patients. Journal of Molecular Diagnostics 22 (3): 405 DOI: 10.1016/j.jmoldx.2019.12.006
[5] Brenner, T.; Decker, S. O.; Grumaz, S.; Stevens, P.; Bruckner, T.; Schmoch, T.; Pletz, M. W.; Bracht, H.; Hofer, S.; Marx, G.; Weigand, M.A.; Sohn, K.; TIFOnet Critical Care Trials Group (2018) Next-generation sequencing diagnostics of bacteremia in sepsis (Next GeneSiS-Trial): Study protocol of a prospective, observational, noninterventional, multicenter, clinical trial, Medicine 97(6): e9868, DOI: 10.1097/MD.0000000000009868
[6] Schmoch, T.; Westhoff, J.H.; Decker, S.O.; Skarabis, A.; Hoffmann, G.F.; Dohna-Schwake, C.; Felderhoff-Müser, U.; Skolik, C.; Feisst, M.; Klose, C.; Bruckner, T.; Luntz, S.; Weigand, M.A.; Sohn, K.; Brenner, T. (2021) Next-generation sequencing diagnostics of bacteremia in pediatric sepsis, Medicine 100(25): e26403, DOI: 10.1097/MD.0000000000026403
 Fraunhofer Institute for Interfacial Engineering and Biotechnology IGB
Fraunhofer Institute for Interfacial Engineering and Biotechnology IGB